Table of Content:
Introduction
- Ever wondered what parameters value to choose as an input for model?
- What should be the maximum depth for decision tree?
- How many trees should be in random forest?
- How many layers should I have in neural network layer or number of neurons in layers?
- What should be the learning rate for model?
And many more questions like this could be answered by Hyperparameter Tuning.
So now the question arises what is Hyperparameter. Basically, it’s a set of parameters which are set to optimize or to get the best performance by model. Now, we don’t know at which values of parameters model will give best result, so we tune the parameters to get optimum result. So we can say that process of finding the combination of hyperparameters which reduces the predefined loss functions and increases the accuracy of given data is known as Hyperparameter Tuning.
Types of Hyperparameter Tuning
We can tune the model by following methods:
- Manual Search
- Grid Search CV
- Randomized Search CV
Manual Search Hyperparameter Tuning
As the name indicates, the process of tuning the hyperparameters manually by trial and error method and by experience is known as Manual Search tuning.
This process is repeated until we don’t get the best hyperparameters which increases the accuracy of model.
Grid Search CV
In Grid Search CV, model is build on every combination of given various hyperparameters and evaluate each model and gives the hyperparameters which have the highest accuracy.
In this technique each set of parameters are considered and model accuracy is noted. The set of parameters which give the highest accuracy is considered to build the model.
Read more about Grid Search in Python here.
Randomized Search CV
In Randomized Search CV, hyperparameters combination are taken randomly to find the best solution for building the model.
As it takes random values, it is faster than grid search cv but gives less accuracy than grid search cv.
Implementation of Randomized Search CV
# Importing Libraries import numpy as np import pandas as pd import matplotlib.pyplot as plt import seaborn as sns import warnings from sklearn.model_selection import train_test_split from sklearn.linear_model import LogisticRegression from sklearn.ensemble import RandomForestRegressor from sklearn.tree import DecisionTreeRegressor from xgboost import XGBRegressor from sklearn.model_selection import RandomizedSearchCV from scipy.stats import randint from xgboost import XGBRegressor from sklearn.metrics import accuracy_score , r2_score from imblearn.over_sampling import SMOTE from sklearn.preprocessing import StandardScaler warnings.filterwarnings('ignore')
# Importing Dataset
# Download red wine quality and white wine quality, sourced from UCI Machine Learning Archive
red = pd.read_csv('winequality-red.csv', sep = ';')
white = pd.read_csv('winequality-white.csv',sep=';')
red.head(5)
fixed acidity volatile acidity citric acid ... sulphates alcohol quality
0 7.4 0.70 0.00 ... 0.56 9.4 5
1 7.8 0.88 0.00 ... 0.68 9.8 5
2 7.8 0.76 0.04 ... 0.65 9.8 5
3 11.2 0.28 0.56 ... 0.58 9.8 6
4 7.4 0.70 0.00 ... 0.56 9.4 5
white.head()
fixed acidity volatile acidity citric acid ... sulphates alcohol quality
0 7.0 0.27 0.36 ... 0.45 8.8 6
1 6.3 0.30 0.34 ... 0.49 9.5 6
2 8.1 0.28 0.40 ... 0.44 10.1 6
3 7.2 0.23 0.32 ... 0.40 9.9 6
4 7.2 0.23 0.32 ... 0.40 9.9 6
# Combining both data
red['Type'] = 'Red'
white['Type'] = 'White'
df = pd.concat([red,white], axis=0)
df.head(5)
fixed acidity volatile acidity citric acid ... alcohol quality Type
0 7.4 0.70 0.00 ... 9.4 5 1
1 7.8 0.88 0.00 ... 9.8 5 1
2 7.8 0.76 0.04 ... 9.8 5 1
3 11.2 0.28 0.56 ... 9.8 6 1
4 7.4 0.70 0.00 ... 9.4 5 1
df.describe()
fixed acidity volatile acidity ... quality Type
count 6497.000000 6497.000000 ... 6497.000000 6497.000000
mean 7.215307 0.339666 ... 5.818378 0.246114
std 1.296434 0.164636 ... 0.873255 0.430779
min 3.800000 0.080000 ... 3.000000 0.000000
25% 6.400000 0.230000 ... 5.000000 0.000000
50% 7.000000 0.290000 ... 6.000000 0.000000
75% 7.700000 0.400000 ... 6.000000 0.000000
max 15.900000 1.580000 ... 9.000000 1.000000
df.info()
Int64Index: 6497 entries, 0 to 4897
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 6497 non-null float64
1 volatile acidity 6497 non-null float64
2 citric acid 6497 non-null float64
3 residual sugar 6497 non-null float64
4 chlorides 6497 non-null float64
5 free sulfur dioxide 6497 non-null float64
6 total sulfur dioxide 6497 non-null float64
7 density 6497 non-null float64
8 pH 6497 non-null float64
9 sulphates 6497 non-null float64
10 alcohol 6497 non-null float64
11 quality 6497 non-null int64
12 Type 6497 non-null int64
dtypes: float64(11), int64(2)
memory usage: 710.6 KB
# check missing values df.isnull().sum() fixed acidity 0 volatile acidity 0 citric acid 0 residual sugar 0 chlorides 0 free sulfur dioxide 0 total sulfur dioxide 0 density 0 pH 0 sulphates 0 alcohol 0 quality 0 Type 0 dtype: int64
df['Type'].value_counts()
white 4898
red 1599
Name: Type, dtype: int64
df['quality'].value_counts()
6 2836
5 2138
7 1079
4 216
8 193
3 30
9 5
Name: quality, dtype: int64
As we can see, the data is imbalanced, we will have to handle this later.
# correlation between features
corr = df.corr(method='spearman')
plt.figure(figsize=(20,10))
sns.heatmap(corr, annot=True)
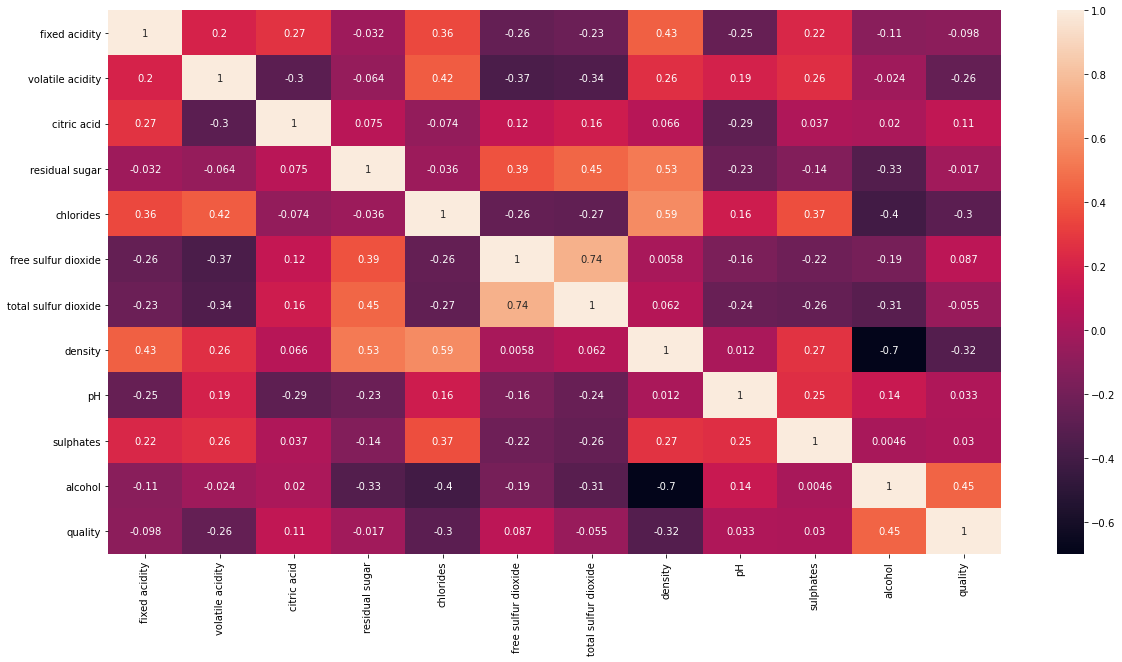
Figure 1 : Correlation plot for selecting best variables
From heatmap in Figure 1, it is clear that the most correlated features with quality are alcohol, density, free sulfur dioxide, chloride and citric acid.
# Splitting the dataset
X = df[['alcohol', 'density', 'free sulfur dioxide', 'chlorides','citric acid']]
y = df['quality']
# Balancing the dataset
oversample = SMOTE(k_neighbors=4)
# transform the dataset
X, y = oversample.fit_resample(X, y)
# Split into train and test sets
X_train,X_test,y_train,y_test = train_test_split(X,y, test_size=0.25, random_state=42)
# Feature Scaling
scaler = StandardScaler()
scaler.fit(X)
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
# Build the Logistic model
lr = LogisticRegression()
lr.fit(X_train,y_train)
lr_pred = lr.predict(X_test)
r2_score(lr_pred,y_test)
0.46038027394992365
# Build the Random Forest Model
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor()
rf.fit(X_train,y_train)
rf_pred = rf.predict(X_test)
r2_score(rf_pred,y_test)
0.916805568147207
# Build Decision Tree Model
dt = DecisionTreeRegressor()
dt.fit(X_train,y_train)
pred_dt = dt.predict(X_test)
r2_score(pred_dt,y_test)
0.8533160326226503
# Build XGBoost Model
xgb = XGBRegressor()
xgb.fit(X_train,y_train)
xg_pred = xgb.predict(X_test)
r2_score(xg_pred,y_test)
0.8705397134271582
Accuracy Score of Random Forest Regressor Model is 91.68%. Now, let’s tune the hyperparameters and see how its effect the accuracy of model.
# Hyperparameter Tuning
rs = {'n_estimators': [100,150,200,300],
'max_features': ['auto', 'sqrt'],
'max_depth': [10, 20, 40, 50, 60, 80, 100, None],
'min_samples_leaf': randint(1,8),
'bootstrap': [True,False]}
random_rf = RandomizedSearchCV(estimator = rf, param_distributions = rs, n_iter = 100, cv = 3, verbose=2, random_state=42, n_jobs = -1)
# Fit the random search model
random_rf.fit(X_train, y_train)
RandomizedSearchCV(cv=3, estimator=RandomForestRegressor(), n_iter=100,
n_jobs=-1,
param_distributions={'bootstrap': [True, False],
'max_depth': [10, 20, 40, 50, 60, 80,
100, None],
'max_features': ['auto', 'sqrt'],
'min_samples_leaf': ,
'n_estimators': [100, 150, 200, 300]},
random_state=42, verbose=2)
random_rf.best_params_
{'bootstrap': False,
'max_depth': 50,
'max_features': 'sqrt',
'min_samples_leaf': 1,
'n_estimators': 300}
random_rf.best_estimator_
RandomForestRegressor(bootstrap=False, max_depth=50, max_features='sqrt', n_estimators=300)
rf1 = RandomForestRegressor(bootstrap=False, ccp_alpha=0.0, criterion='mse',
max_depth=50, max_features='sqrt', max_leaf_nodes=None,
max_samples=None, min_impurity_decrease=0.0,
min_impurity_split=None, min_samples_leaf=1,
min_samples_split=2, min_weight_fraction_leaf=0.0,
n_estimators=300, n_jobs=None, oob_score=False,
random_state=None, verbose=0, warm_start=False)
rf1.fit(X_train,y_train)
RandomForestRegressor(bootstrap=False, max_depth=50, max_features='sqrt',
n_estimators=300)
rf_pr = rf1.predict(X_test)
r2_score(rf_pr,y_test)
0.9322745287584387
Conclusion
With the help of hyperparameter tuning models accuracy increased by 2% and now it is 93.22%.
Randomized Search CV is faster than Grid Search CV. So, for big data we can use Randomized Search CV instead of Grid Search but if higher accuracy is needed than we should go with Grid search.
About the Author's:

Write A Public Review